
The EBI has a new phylogeny-aware multiple sequence alignment program which makes use of evolutionary information to help place insertions and deletions. Transform a Sequence Similarity Search result into a Multiple Sequence Alignment or reformat a Multiple Sequence Alignment using the MView program.Ĭonsistency-based MSA tool that attempts to mitigate the pitfalls of progressive alignment methods. Suitable for medium-large alignments.Īccurate MSA tool, especially good with proteins. MSA tool that uses Fast Fourier Transforms. Very fast MSA tool that concentrates on local regions. Suitable for medium-large alignments.ĮMBOSS Cons creates a consensus sequence from a protein or nucleotide multiple alignment. New MSA tool that uses seeded guide trees and HMM profile-profile techniques to generate alignments.
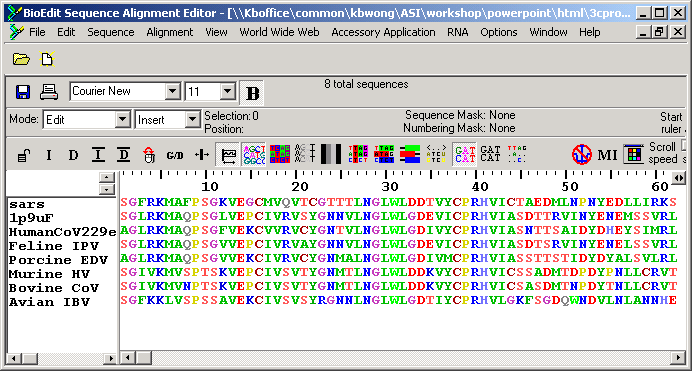
From the output, homology can be inferred and the evolutionary relationships between the sequences studied.īy contrast, Pairwise Sequence Alignment tools are used to identify regions of similarity that may indicate functional, structural and/or evolutionary relationships between two biological sequences. Multiple Sequence Alignment (MSA) is generally the alignment of three or more biological sequences (protein or nucleic acid) of similar length.


 0 kommentar(er)
0 kommentar(er)
